Fri Dec 26 00:30:00 UTC 2025: Here’s a summary of the text:
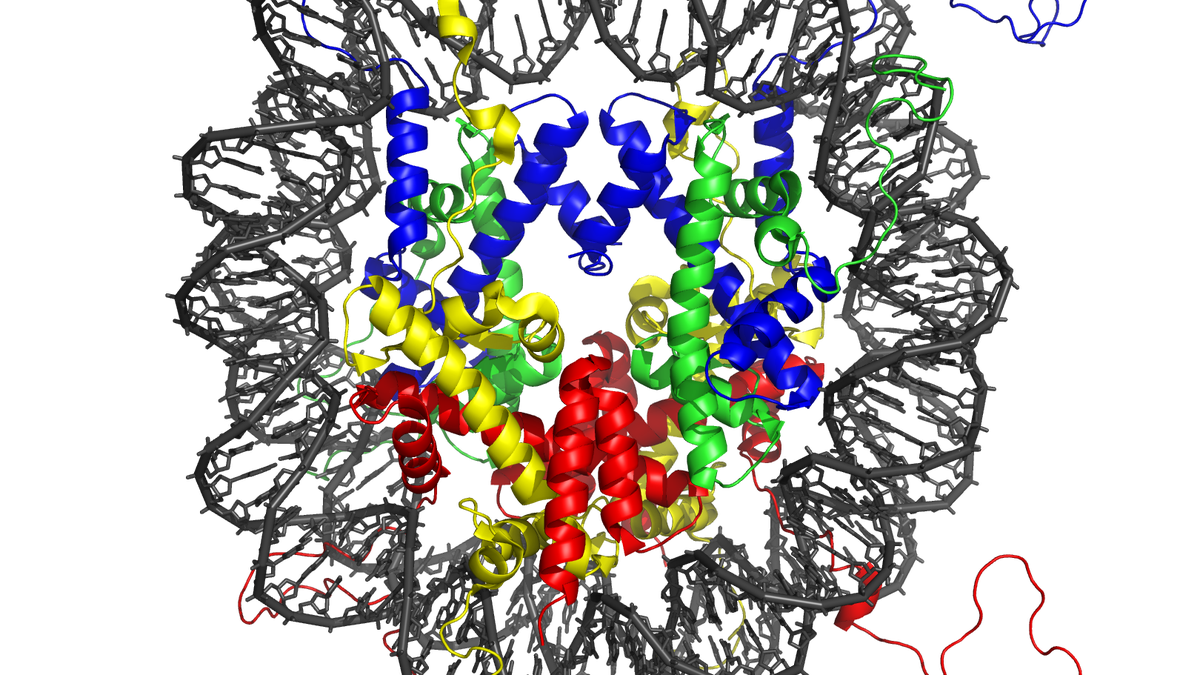
A new study published in Science reveals that the spacing between DNA-protein units (nucleosomes) in chromatin, the structure that packages DNA within cells, significantly impacts chromatin behavior. Even small variations in this spacing alter the orientation of the nucleosomes, influencing how chromatin interacts with neighboring strands and affecting its overall structure and stability. Researchers built chromatin in the lab with varying linker DNA lengths and observed that shorter linkers resulted in densely connected, rigid structures, while longer linkers produced more fluid, less stable structures. This difference in structure is thought to impact gene accessibility and regulation. Experts believe these findings provide insights into understanding genome instability and potential links between chromatin’s physical state and gene regulation across different cell types.
Here’s a news article based on the text:
Tiny DNA Spacing, Huge Impact: New Study Reveals Key to Chromatin Behavior
The Hindu, New Delhi – December 26, 2025 – A groundbreaking study published in Science has unveiled a surprisingly critical factor in understanding how DNA is organized and regulated within cells: the spacing between DNA-protein units called nucleosomes within chromatin.
Researchers at UT Southwestern Medical Center, led by Professor Michael Rosen, discovered that even slight variations in the length of the “linker DNA” connecting these nucleosomes can dramatically alter the overall structure and behavior of chromatin. Chromatin, a complex of DNA and proteins, is vital for packaging the body’s genetic material into the tiny space of a cell nucleus. Its arrangement also influences which genes are accessible for reading and which are shut down.
The team created artificial chromatin in the lab with differing linker DNA lengths and found that chromatin with shorter linkers formed dense, rigid structures that readily tangled, while chromatin with longer linkers created more fluid, less stable structures. These structural differences can affect how easily regulatory molecules move through and access the DNA.
“Those different interaction patterns are what make one system behave like a simple liquid and the other behave more like silly putty or toothpaste,” explained Professor Rosen.
Experts believe the findings could have significant implications for understanding genome instability in diseases like cancer and aging, particularly in repetitive DNA regions. Furthermore, the study raises the possibility that chromatin’s physical state could influence how genes are regulated across different cell types.
“The genome’s organisation is encoded in the chromatin itself. You don’t need additional instructions to make structure emerge.” said National Institutes of Health biochemist Yamini Dalal.
Sarah Teichmann, a professor at Cambridge University, noted that large-scale projects like the Human Cell Atlas could utilize these findings to explore whether such physical chromatin states vary with cell identity.
While further research is needed to fully understand the extent to which cells actively use this spacing to regulate chromatin function, the study provides a crucial physical blueprint for understanding genome organization and function.
